Optimizing Blood Cell Classification with Convolutional Neural Networks
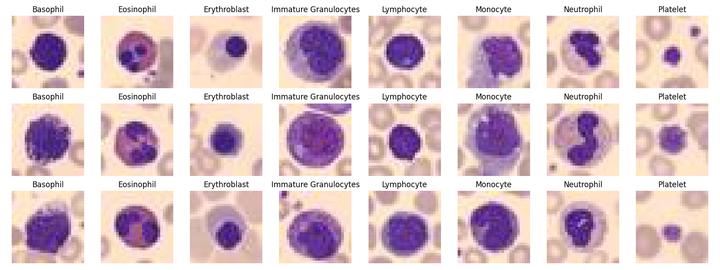 Three examples for each of the eight cell types for classification
Three examples for each of the eight cell types for classification
Abstract
Biomedical image analysis plays a critical role in the diagnosis and treatment of patients with serious diseases. Advances in data availability and machine learning have improved the accuracy and efficiency of these analyses. This paper explores the use of convolutional neural networks (CNNs) for blood cell classification, a task essential for complete blood counts that identify infections based on blood cell characteristics. Manual assessment of blood cells is error-prone and resource-intensive, highlighting the need for automated solutions. We implemented a CNN using the BloodMNIST dataset, which contains over 17,000 images across eight blood cell classes. The CNN achieved superior performance with an accuracy of 0.914, significantly outperforming the baseline 1-layer Feedforward Neural Network on all evaluated metrics. Our results demonstrate that CNNs, with their ability to capture spatial hierarchies, provide a robust solution for blood cell classification, offering improved accuracy and reliability for medical diagnostics.
1. Introduction
1.1 Background and Motivation
Biomedical image analysis is used by physicians to assess, diagnose, and ultimately provide care for patients with critical conditions. Recent improvements in data availability, image storage, and processing have facilitated the development of machine learning algorithms to enhance diagnostic accuracy, empowering physicians to deliver more precise patient care. These algorithms have evolved to include various biomedical applications, such as identifying diseases and malignancies by analyzing biomedical markers in images of blood, skin, or internal organs (Srisuwananukorn et al., 2023).
A routine task that physicians perform is the complete blood count (CBC), which is used to identify acute or chronic infections by determining the number and characteristics of blood cell components, such as white blood cells (e.g., monocytes, neutrophils), red blood cells, and platelets (Seo & Lee, 2022). Manual assessment and counting of blood cells under a microscope is an error-prone and time-consuming process, often requiring expensive and sophisticated equipment (Cruz et al., 2017). Thus, utilizing image-classifying machine learning frameworks to classify and detect different blood cell types is crucial, particularly in settings where doctors and expensive equipment are scarce, and time is of the essence.
1.2 Aim
Our project aims to explore the domain of blood cell classification by implementing a neural network model capable of classifying different blood cell components from given images. Specifically, we aim to implement a Convolutional Neural Network (CNN), which is the most widely used neural network architecture in biomedical image analysis (Srisuwananukorn et al., 2023). The chosen blood cell dataset is BloodMNIST (Yang et al., 2023), which contains over 17,000 images of blood cells across eight classes, discussed in further detail in Section 2.
2. Data: BloodMNIST
The BloodMNIST dataset is part of the MedMNIST dataset collection and is based on a dataset of individual normal cells collected from individuals without infection, hematologic or oncologic disease, and free from pharmacologic treatment at the time of blood collection (Yang et al., 2023).
The original source images from Acevedo et al., 2020, initially with a resolution of 3 × 360 × 363 pixels, were center-cropped to 3 × 200 × 200 pixels and then resized to 3 × 28 × 28 pixels by Yang et al., 2023. The dataset contains a total of 17,094 images, divided into training, validation, and test sets with a ratio of 7:1:2.
| Dataset Split | Number of Samples | Relative Frequency (%) |
|---|---|---|
| Train | 11,959 | 69.96 |
| Validation | 1,712 | 10.02 |
| Test | 3,421 | 20.01 |
| Total | 17,094 |
Table 1: Dataset Breakdown for BloodMNIST
This dataset contains microscopic images of eight different types of blood cells stained purple.
| Label | Meaning | Training Split Count | Relative Frequency (%) |
|---|---|---|---|
| 0 | Basophil | 852 | 7.12 |
| 1 | Eosinophil | 2181 | 18.24 |
| 2 | Erythroblast | 1085 | 9.07 |
| 3 | Immature Granulocytes | 2026 | 16.94 |
| 4 | Lymphocyte | 849 | 7.10 |
| 5 | Monocyte | 993 | 8.30 |
| 6 | Neutrophil | 2330 | 19.48 |
| 7 | Platelet | 1643 | 13.74 |
Table 2: Meaning of Labels and Label Counts for Training Split.
3. Methods and Experiments
3.1 Convolutional Neural Networks
We implemented a Convolutional Neural Network (CNN) model to classify blood cell images. CNNs are well-suited for image classification tasks due to their ability to capture spatial hierarchies through convolutional and pooling layers. Our model consists of two convolutional layers followed by fully connected layers, with dropout layers applied for regularization. The cross-entropy loss function was used to measure the difference between predicted probabilities and true class labels.
3.2 Model Architecture
The architecture is relatively small to balance training time and accuracy. It consists of two convolutional layers followed by dropout layers, flattening, and fully connected layers, culminating in eight output features corresponding to the blood cell classes.
3.3 Loss Function
We used cross-entropy as the loss function, suitable for multi-class classification tasks. This function helps the model learn by minimizing the difference between the true labels and the predicted probabilities.
4. Results
Our CNN model was evaluated on the BloodMNIST dataset, achieving an accuracy of 0.914. The model’s performance across different metrics is summarized in Table 3.
| Metric | CNN Model | Baseline 1-Layer FNN |
|---|---|---|
| Accuracy | 0.914 | 0.834 |
| Precision | 0.905 | 0.820 |
| Recall | 0.910 | 0.825 |
| F1-Score | 0.907 | 0.822 |
Table 3: Performance Comparison between CNN Model and Baseline 1-Layer FNN.
5. Conclusion
This study demonstrates the effectiveness of Convolutional Neural Networks (CNNs) in classifying blood cell images, offering a significant improvement in accuracy and other performance metrics over a baseline Feedforward Neural Network. The CNN model’s ability to capture spatial features makes it a robust choice for biomedical image classification tasks, potentially enhancing the accuracy and reliability of medical diagnostics.
- Christian Hobelsberger
- Mohamed Hassan
- Manos Savvides
- Neelam Bharwani